Keywords: web scraping, file parsing, PHP programming, regular expressions, MHC-peptide binding, Immunoinformatics
Logical flow of the T-Score application
This web application will accept a protein sequence or UniProt id as input. The user will be able to select an HLA (MHC) allele for which the user wishes to calculate the binding score to all the possible 9mers (sequences of 9 amino-acids) present in the input sequence and the threshold above which positive peptides will be shown in the analysis results.
The processing script will generate all possible 9mers from the input protein and assign each one a binding score to the selected HLA allele, based on the appropriate scoring matrix, as discussed in the previous section. For each peptide the score will be used to rank the peptide according to the thresholds present in the scoring matrix, thereby assigning the peptide to say, the top 10% binders, the top 5% binders, the top 1% binders etc.
An output will be generated that will list, for each protein, a list of the peptides whose score ranks them above the threshold selected by the user in the initial web form.
As you can imagine, there is quite some code to be written to achieve all these functionalities.
In this section we will concentrate on gathering the scoring matrices data from the original web page where they are shared, through a web scraping operation performed by using PHP and regular expressions.
Downloading the scoring matrices raw HTML files from the original website and saving a local copy
As a first step, we need to download the data relative to each scoring matrix from the website where they are available. The operation of extracting data we need from an existing web page is called “web scraping”. Web scraping can be achieved by different techniques. For this example we will adopt a simple strategy based on the manual analysis of the HTML source code of our target pages. From this analysis we will be able to write dedicated regular expressions and use those in an appropriate code context so as to retrieve the scoring matrix data and write it to text files.
Another piece of code will be dedicated to reading those text files and storing the data into appropriate data structures (arrays) that can then be used by the form processing script to assign scores to peptides.
Let’s get started.
Our application will be limited to the analysis with three HLA alleles, namely HLA-A1, A2 and A3. The scoring matrices for these three alleles can be found at the following URLs:
A1 matrix: http://www.imtech.res.in/raghava/nhlapred/matrices/a1.html
A2 matrix: http://www.imtech.res.in/raghava/nhlapred/matrices/a2.html
A3 matrix: http://www.imtech.res.in/raghava/nhlapred/matrices/a2.html
Let us create somewhere within the path of the web root of our apache installation, a directory for the whole project, called tscore and, inside, a subdirectory called matrix, where the matrix work will be executed and the actual matrices files will be stored. The code will be written in a file called “script.php” to be created inside the tscore directory. To avoid permissions issues during code execution, grant a 777 permission to both the tscore and matrix directories with chmod (chmod 777 directory_path). So for now we will have the following structure:
tscore (permission 777)
matrix (permission 777)
index.php
Into script.php the very first task will be to download the three matrices “raw” HTML files, as they are, into the matrix folder, if they do not exist in this folder already (we do not want to create any unnecessary load to the server from which we are scraping the data, so let us ensure we download the original html pages just once).
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 |
<?php $urls = array("http://www.imtech.res.in/raghava/nhlapred/matrices/a1.html", "http://www.imtech.res.in/raghava/nhlapred/matrices/a2.html", "http://www.imtech.res.in/raghava/nhlapred/matrices/a3.html"); foreach($urls as $url){ preg_match("/a\d\.html/", $url, $matches); // We are looking for the final "ax.html" part of the URL $page_name = $matches[0]; // and store it into a variable called $page_name $local_page_path = "matrix/$page_name"; // We will store the files with their original names in the matrix folder, here we define the relative path with respect to script.php if(!file_exists($local_page_path)){ // We check that the file is not there already, and if not $handle = fopen($local_page_path, "w") or die("Could not create the file"); // we create an handle for it, in write mode "w". // If the handle cannot be created we will get an error message and the script will abort, thanks to the die command // if you get such an error check the directories permissions $raw_html_file = file_get_contents($url); // we download it from the web site and store it's contents in a variable fwrite($handle, $raw_html_file) or die("Could not write to the file"); // then write the variable contents to the local file // If the file cannot be written to we will get an error message and the script will abort, thanks to the die command // if you get such an error check the directories permissions fclose($handle); // and close the file handle } } ?> |
You can try the code above yourself and then check that the expected 3 files a1.html, a2.html and a3.html were created in the matrix folder and do contain some HTML code. Thank you for doing this just once (if successful), so as to avoid any unnecessary load on the server thanks to which the matrices are kindly shared by the research group who created them.
It is worth mentioning that instead of using the fopen(), fwrite() and fclose() funtions sequentially, as we did in the code above, we could have used the file_put_contents() function, which can be considered a shortcut to these three functions for file writing. We have used the long way to do it for educational purposes here.
So we could replace this part of the code above:
|
1 2 3 4 5 6 7 8 9 10 |
$handle = fopen($local_page_path, "w") or die("Could not create the file"); // we create an handle for it, in write mode "w". // If the handle cannot be created we will get an error message and the script will abort, thanks to the die command, // if you get such an error check the directories permissions $raw_html_file = file_get_contents($url); // we download it from the web site and store it's contents in a variable fwrite($handle, $raw_html_file) or die("Could not write to the file"); // then write the variable contents to the local file // If the file cannot be written to we will get an error message and the script will abort, thanks to the die command, // if you get such an error check the directories permissions fclose($handle); // and close the file handle |
with something much shorter, like:
|
1 2 3 |
file_put_contents($local_page_path, file_get_contents($url)) or die("something went wrong"); |
Scraping the scoring matrices data from the HTML files
We can now proceed to extract the data from those HTML files and write the data, in a cleaner format, in some new text files that we will name as a1-matrix.csv, a2-matrix.csv and a3-matrix.csv.
The problem we have is that in the original html files the various numbers and amino-acids letters we need are included into an HTML table, so they are heavily mixed-up with HTML tags that we need to get rid of. Click on the image below to get an idea:

Let’s state upfront where we are aiming at. We want to generate a file with the matrix data in csv format, where the scoring values for each amino-acid are stored in a dedicated line and separated by semicolons. The first character of each line will be the letter of the amino-acid to which the numbers that follow refer to. So each file will contain 21 lines (one for each amino-acid plus the “generic” amino-acid X, which is included in the matrices). Each one of these lines will contain the amino-acid letter as first character followed by 9 numbers, corresponding to the numeric scoring values for the P1 to P9 positions.
To clarify the concept, let’s consider again the scoring matrix for the HLA-A1 allele:
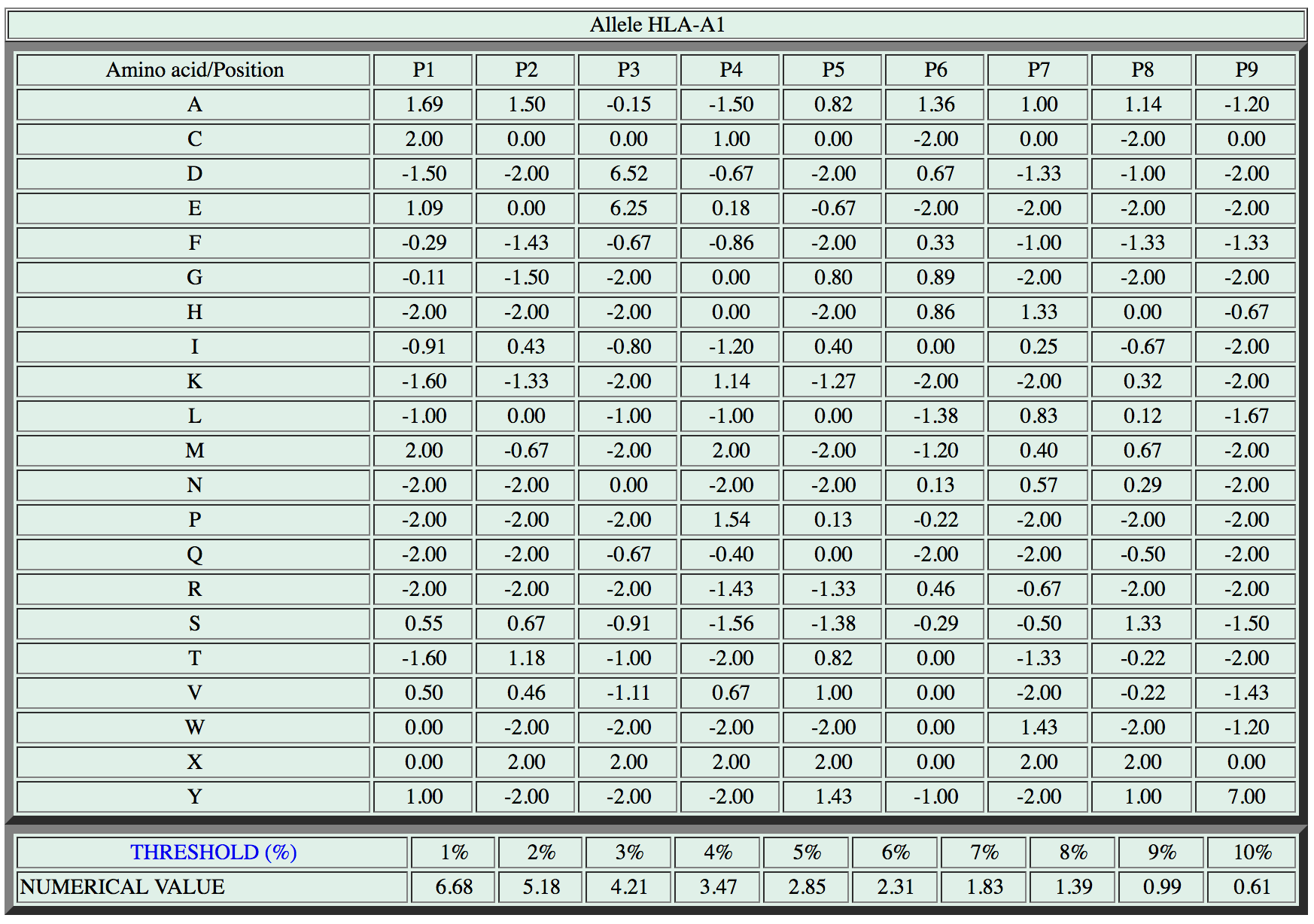
Our text file for the HLA-A1 matrix (a1-matrix.csv) will start with those two lines:
A;1.69;1.50;-0.15;-1.5;0.82;1.36;1.00;1.14;-1.20
C;2.00;0.00;0.00;1.00;0.00;-2.00;0.00;-2;00;0.00
etc…..
In the same file we will also store the threshold information (last two line in the matrix figure above), but let’s take care about this later on.
In order to extract the data from the HTML file (web scraping) we need to carefully look at the HTML source code of the source page(s). Please open this page in your browser, the HTML page for the HLA-A1 matrix, and look at the source HTML code by selecting the appropriate option in the browser’s menu.
At the time of this writing, this is what the relevant part of the HTML, the one with the matrix data, looks like. You can click on the image to view a larger version. Even better, look at the original source code in your browser.

Thankfully, all the lines for each amino-acid look the same in the source code and each amino-acid occupies it’s own line which, in HTML terms, is a table raw, embedded in TR tags (capital letters as this page is written in “old style” HTML1). Let’s consider the line for the first amino-acid Alanine (A):
|
1 2 3 |
<TR><TD><CENTER>A</CENTER></TD><TD><CENTER>1.69</CENTER></TD><TD><CENTER>1.50</CENTER></TD><TD><CENTER>-0.15</CENTER></TD><TD><CENTER>-1.50</CENTER></TD><TD><CENTER>0.82</CENTER></TD><TD><CENTER>1.36</CENTER></TD><TD><CENTER>1.00</CENTER></TD><TD><CENTER>1.14</CENTER></TD><TD><CENTER>-1.20</CENTER></TD></TR> |
See how all the data we need is there, we just need to scrape it out.
We start by writing a regular expression that matches the line with a capture group right where we need it.
More specifically, we are interested in matching parts like this:
|
1 2 3 |
<TD><CENTER>A</CENTER></TD> |
like this:
|
1 2 3 |
<TD><CENTER>1.69</CENTER></TD> |
or like this:
|
1 2 3 |
<TD><CENTER>-0.15</CENTER></TD> |
So our capture group may contain from 1 to 5 characters.
Here is a fitting expression:
|
1 2 3 |
$regexp = "/<TD><CENTER>(.{1,5})<\/CENTER><\/TD>/"; |
We can then use preg_match_all() to capture all the matches, in the line, to the capture group. The matches will be either a capital letter (the amino-acid letter, should be the first match found), or score numbers.
Before we get into the parsing of a whole file, let’s try the regular expression on a single line with a small example:
|
1 2 3 4 5 6 7 8 9 |
<?php $str = "<TR><TD><CENTER>A</CENTER></TD><TD><CENTER>1.69</CENTER></TD><TD><CENTER>1.50</CENTER></TD><TD><CENTER>-0.15</CENTER></TD><TD><CENTER>-1.50</CENTER></TD><TD><CENTER>0.82</CENTER></TD><TD><CENTER>1.36</CENTER></TD><TD><CENTER>1.00</CENTER></TD><TD><CENTER>1.14</CENTER></TD><TD><CENTER>-1.20</CENTER></TD></TR>"; $regexp = "/TD><CENTER>(.{1,5})<\/CENTER><\/TD>/"; preg_match_all($regexp, $str, $matches); var_dump($matches[1]); ?> |
This will output the following var_dump:
array(10) {
[0]=>
string(1) “A”
[1]=>
string(4) “1.69”
[2]=>
string(4) “1.50”
[3]=>
string(5) “-0.15”
[4]=>
string(5) “-1.50”
[5]=>
string(4) “0.82”
[6]=>
string(4) “1.36”
[7]=>
string(4) “1.00”
[8]=>
string(4) “1.14”
[9]=>
string(5) “-1.20”
}
This is great as we now have an array, namely $matches[1] (remember that $matches[0] contains the matches to the whole regular expression while we need the matches for the capture group, that are indeed in $matches[1]), that contains exactly the data we were looking for.
Let’s now discuss the thresholds part. The page(s) we are parsing to scrape the data actually contains six different tables. The fifth table contains the scoring data we just discussed while the sixth table contains the thresholds data. Check this out in the HTML source of one of the original pages.
We need to manage this in the script. In particular, as we reach the sixth table, the thresholds one, during the parsing process, we know we should stop scraping scoring data and instead start to scrape the thresholds data.
To keep track about which table we are managing at a given time into the scraping code, we will use a “$table_flag” variable whose value will be 0 to begin with, will become 5 as we reach the scores table and 6 as we reach the thresholds table. Depending on the value of $table_flag we will execute a different code.
In the .csv file, thresholds will be represented by a dedicated line (the last line of the file) with this format:
tre;6.68;5.18;4.21;3.47;2.85;2.31;1.83;1.39;0.99;0.61 (this is how the last line of the .csv file for the HLA-A1 matrix will look like)
We can now parse the files. To do this, we will list all the files in the matrix directory. Then, file name by file name, if the file name ends in .html and the respective .csv file does not exist, we will create it and put inside the cleaned matrix scoring and thresholds data, in the csv format we discussed above.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 |
<?php $file_names = scandir("matrix"); // An array with all the names of the files in the matrix directory $score_regexp = "/<TD><CENTER>(.{1,5})<\/CENTER><\/TD>/"; $threshold_regexp = "/<td><center>(.{4})<\/center><\/td>/"; // Unlike those for the score values, html tags for the threshold values are in lowercase in the original pages we are scraping the data from foreach($file_names as $file_name){ // Iterating through the files, one by one if(preg_match("/^(.+)\.html$/", $file_name, $matches) and !file_exists("matrix/".$matches[1]."-matrix.csv")){ $html_file_path = "matrix/$file_name"; $csv_file_path = "matrix/".$matches[1]."-matrix.csv"; $html_file_contents = file_get_contents($html_file_path); $html_file_lines = explode("\n", $html_file_contents); $csv_handle = fopen($csv_file_path, "w"); $table_flag = 0; // Each time we come across an opening table tag during file parsing, this will be raised by one. $threshold_csv_line_temp = "\ntre;"; $j = 1; // Counter for the thresholds lines // File parsing to scrape the data may begin! foreach($html_file_lines as $line){ $line = trim($line); // You never know which weird characters may be lurking at the beginning or end of your lines... if(preg_match("/<table/", $line)){ $table_flag++; } if($table_flag == 5){ // We are parsing one of the lines belonging the scores table if(preg_match_all($score_regexp, $line, $matches) and !preg_match("/P1/", $line)){ $i = 1; foreach($matches[1] as $match){ if($i < 10){ fwrite($csv_handle, $match.";"); // We want to add a semicolon after each of the element but the last one and we have a total of 10 elements, one aminoacid letter plus 9 scores $i++; } elseif($i == 10){ fwrite($csv_handle, $match."\n"); // After the last score we add a newline instead of a semicolon } } } } elseif($table_flag == 6){ // We are parsing one of the lines belonging the thresholds table if(preg_match($threshold_regexp, $line, $matches)){ // We use preg_match instead of preg_match_all as in the original files HTML source each threshold is in it's own line so we just need to grab one value here // we also manage things a little differently, adding the values to a variable instead of writing them to the file directly, for the same reason. if($j < 10){ $threshold_csv_line_temp = $threshold_csv_line_temp.$matches[1].";"; $j++; } elseif($j == 10){ // The last threshold value $threshold_csv_line_temp = $threshold_csv_line_temp.$matches[1]; } } } } fwrite($csv_handle, $threshold_csv_line_temp); fclose($csv_handle); } } ?> |
Mind that the execution of this last bit of code assumes that the html files were already downloaded to the matrix directory by executing the download code upper in the page. After running this code the matrix directory will contain 3 new files: a1-matrix.csv, a2-matrix.csv and a3-matrix.csv. Here is what the a1-matrix.csv file contents will look like:
A;1.69;1.50;-0.15;-1.50;0.82;1.36;1.00;1.14;-1.20
C;2.00;0.00;0.00;1.00;0.00;-2.00;0.00;-2.00;0.00
D;-1.50;-2.00;6.52;-0.67;-2.00;0.67;-1.33;-1.00;-2.00
E;1.09;0.00;6.25;0.18;-0.67;-2.00;-2.00;-2.00;-2.00
F;-0.29;-1.43;-0.67;-0.86;-2.00;0.33;-1.00;-1.33;-1.33
G;-0.11;-1.50;-2.00;0.00;0.80;0.89;-2.00;-2.00;-2.00
H;-2.00;-2.00;-2.00;0.00;-2.00;0.86;1.33;0.00;-0.67
I;-0.91;0.43;-0.80;-1.20;0.40;0.00;0.25;-0.67;-2.00
K;-1.60;-1.33;-2.00;1.14;-1.27;-2.00;-2.00;0.32;-2.00
L;-1.00;0.00;-1.00;-1.00;0.00;-1.38;0.83;0.12;-1.67
M;2.00;-0.67;-2.00;2.00;-2.00;-1.20;0.40;0.67;-2.00
N;-2.00;-2.00;0.00;-2.00;-2.00;0.13;0.57;0.29;-2.00
P;-2.00;-2.00;-2.00;1.54;0.13;-0.22;-2.00;-2.00;-2.00
Q;-2.00;-2.00;-0.67;-0.40;0.00;-2.00;-2.00;-0.50;-2.00
R;-2.00;-2.00;-2.00;-1.43;-1.33;0.46;-0.67;-2.00;-2.00
S;0.55;0.67;-0.91;-1.56;-1.38;-0.29;-0.50;1.33;-1.50
T;-1.60;1.18;-1.00;-2.00;0.82;0.00;-1.33;-0.22;-2.00
V;0.50;0.46;-1.11;0.67;1.00;0.00;-2.00;-0.22;-1.43
W;0.00;-2.00;-2.00;-2.00;-2.00;0.00;1.43;-2.00;-1.20
X;0.00;2.00;2.00;2.00;2.00;0.00;2.00;2.00;0.00
Y;1.00;-2.00;-2.00;-2.00;1.43;-1.00;-2.00;1.00;7.00
tre;6.68;5.18;4.21;3.47;2.85;2.31;1.83;1.39;0.99;0.61
Let’s put all the code for this section together, in a single script that will download the HTML files from the matrices web site and extract the data to .csv files in a split second:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 |
<?php $urls = array("http://www.imtech.res.in/raghava/nhlapred/matrices/a1.html", "http://www.imtech.res.in/raghava/nhlapred/matrices/a2.html", "http://www.imtech.res.in/raghava/nhlapred/matrices/a3.html"); foreach($urls as $url){ preg_match("/a\d\.html/", $url, $matches); // We are looking for the final "ax.html" part of the URL $page_name = $matches[0]; // and store it into a variable called $page_name $local_page_path = "matrix/$page_name"; // We will store the files with their original names in the matrix folder, here we define the relative path with respect to script.php if(!file_exists($local_page_path)){ // We check that the file is not there already, and if not file_put_contents($local_page_path, file_get_contents($url)) or die("something went wrong"); } } $file_names = scandir("matrix"); // An array with all the names of the files in the matrix directory $score_regexp = "/<TD><CENTER>(.{1,5})<\/CENTER><\/TD>/"; $threshold_regexp = "/<td><center>(.{4})<\/center><\/td>/"; // Unlike those for the score values, html tags for the threshold values are in lowercase in the original pages we are scraping the data from foreach($file_names as $file_name){ // Iterating through the files, one by one if(preg_match("/^(.+)\.html$/", $file_name, $matches) and !file_exists("matrix/".$matches[1]."-matrix.csv")){ $html_file_path = "matrix/$file_name"; $csv_file_path = "matrix/".$matches[1]."-matrix.csv"; $html_file_contents = file_get_contents($html_file_path); $html_file_lines = explode("\n", $html_file_contents); $csv_handle = fopen($csv_file_path, "w"); $table_flag = 0; // Each time we come across an opening table tag during file parsing, this will be raised by one. $threshold_csv_line_temp = "\ntre;"; $j = 1; // Counter for the thresholds lines // File parsing to scrape the data may begin! foreach($html_file_lines as $line){ $line = trim($line); // You never know which weird characters may be lurking at the beginning or end of your lines... if(preg_match("/<table/", $line)){ $table_flag++; } if($table_flag == 5){ // We are parsing one of the lines belonging the scores table if(preg_match_all($score_regexp, $line, $matches) and !preg_match("/P1/", $line)){ $i = 1; foreach($matches[1] as $match){ if($i < 10){ fwrite($csv_handle, $match.";"); // We want to add a semicolon after each of the element but the last one and we have a total of 10 elements, one aminoacid letter plus 9 scores $i++; } elseif($i == 10){ fwrite($csv_handle, $match."\n"); // After the last score we add a newline instead of a semicolon } } } } elseif($table_flag == 6){ // We are parsing one of the lines belonging the thresholds table if(preg_match($threshold_regexp, $line, $matches)){ // We use preg_match instead of preg_match_all as in the original files HTML source each threshold is in it's own line so we just need to grab one value here // we also manage things a little differently, adding the values to a variable instead of writing them to the file directly, for the same reason. if($j < 10){ $threshold_csv_line_temp = $threshold_csv_line_temp.$matches[1].";"; $j++; } elseif($j == 10){ // The last threshold value $threshold_csv_line_temp = $threshold_csv_line_temp.$matches[1]; } } } } fwrite($csv_handle, $threshold_csv_line_temp); fclose($csv_handle); } } ?> |
There you have it, a file for each one of the three selected MHC alleles with all the data needed to score a peptide for binding, in a clean format. In the next section we will reason on how to use these file to actually score a peptide and then rank it according to the available thresholds.
Chapter Sections
[pagelist include=”1461″]
[siblings]
WORK IN PROGRESS ON CHAPTER 5!